Epigenetics Tutorial
Introduction
Epigenetics is a relatively new field born from two parent sciences: genetics and developmental biology. [1]Holliday, R. Epigenetics: A historical overview. Epigenetics 1, 76–80 (2006). The prefix “epi” means “above” or “in addition to.” [2]Harper, D. Online Etymology Dictionary. (2014). at http://www.etymonline.com In order to understand what epigenetics is “in addition to,” or what it is “above,” we must meet its parent sciences.
Developmental biology is the study of how organisms grow and develop across their lives, from conception through the many changes that come with advanced age (senescence).
Developmental biology is the study of how organisms grow and develop across their lives, from conception through the many changes that come with advanced age (senescence).

Multicellular organisms, such as human beings, begin as single cells. These single cells multiply and diverge from each other many times to form tissues and organs with unique characteristics.
These tissues and organs work together to respond to the environment, and to each other, in a vast and complex system. In order for an organism to survive, this system must constantly work to repair itself and maintain equilibrium, while remaining flexible enough to endure or adapt to fluctuations introduced by its surroundings. Certain environmental conditions introduced at key times or over a long duration can have dramatic effects on the biological development of human beings.
Genetics is the study of the biological material called deoxyribonucleic acid (DNA), its contents, its structure, its function, and its transmission across generations through heredity.

We do not yet understand the function of all our DNA. Some parts of our DNA act as a repository for genes. Genes signal the body to make certain compounds (usually proteins) in a highly coordinated fashion. Other parts of our DNA contain directions about how and when to read the instructions called genes. The sum total of an organism’s DNA, including all of its genes, is called a genome. Although all parts of the body (usually) contain the same genome, specialized tissues and organs have restricted access to different parts of the genome at different times. For example, genes solely related to the function of the skeletal system are accessed by the cells that make up bones and not by the cells that make up the heart.
Epigenetics is the study of changes above the level of the genome. These changes do not affect the underlying content of the DNA; the recipes encoded by the genes remain the same. Instead, epigenetic changes alter the genome’s superstructure, or its architecture, in a way that impacts how the genome functions. By altering the genomic architecture, epigenetic changes can restrict a cell’s access to just those genes that are relevant to the cell’s purpose in the body. Epigenetics explains how cells in disparate tissues and organs can have the same genome, but look and act completely differently. By making small adjustments to the architecture of a genome, epigenetic changes give human beings the flexibility they need to develop from a single cell to a complex multi-cellular organism, and to continue responding to a dynamic environment across the lifespan.
Human DNA is (with few exceptions) not “edited” by one’s environment. The epigenome, on the other hand, is responsive to environmental changes across a lifetime. Changes in the epigenome are a normal part of life course development, but they have also been associated with certain health problems, such as type 2 diabetes[3]Pinney, S. E. & Simmons, R. a. Epigenetic mechanisms in the development of type 2 diabetes. Trends Endocrinol. Metab. 21, 223–9 (2010). and cancer.[4]Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–59 (2008). Some epigenetic changes can even be passed down from parents to their children.

Historically, there has been debate over the official definition of “epigenetics.” Most of this debate has centered on the issue of heritability. Cold Spring Harbor and the NIH Roadmap Epigenomics Mapping Consortium represent two respected institutions whose definitions of epigenetics are at odds with each other:
An epigenetic trait is a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence.[5]Berger, S. L., Kouzarides, T., Shiekhattar, R. & Shilatifard, A. An operational definition of epigenetics. Genes Dev. 23, 781–3 (2009).
[…] epigenetics refers to both heritable changes in gene activity and expression (in the progeny of cells or of individuals) and also stable, long-term alterations in the transcriptional potential of a cell that are not necessarily heritable.[6]NIH Roadmap Epigenomics. Roadmap Epigenomics Project – Overview. (2010). at http://www.roadmapepigenomics.org/overview.
Stricter definitions of epigenetics, like Cold Spring Harbor’s, require epigenetic changes to be heritable across cell divisions (mitotically), across generations of an organism (meiotically), or both. More inclusive definitions of epigenetics, like the NIH Roadmap Epigenomics Mapping Consortium’s, do not require modifications to be heritable, but suggest that they should have long-term effects on gene expression (when a gene is “turned on” and to what degree). The argument over heritable vs. non-heritable changes is important because its resolution determines which types of modifications are ultimately considered “epigenetic” and which fall into the more general category of “biochemical regulation.” For the purpose of this tutorial, we will rely on the more liberal definition provided by the NIH Roadmap Epigenomics Mapping Consortium. The consequence is that not all scientists will agree that each of the mechanisms described in this tutorial is necessarily “epigenetic.” Throughout the tutorial, we will use a note to mark wherever different definitions of epigenetics might cause a conflict in interpretation.
Epigenetics has built bridges between the fields of genetics and developmental biology, and has continued to grow new connections across disciplines, including psychology, epidemiology, neuroscience, nutrition, and many others. As the line between disciplines continues to blur, epigenetics is an increasingly useful tool for both bioscientists and social scientists. To gain a deeper understanding of epigenetic mechanisms and their impact on the human condition, a working knowledge of gene expression and genome structure are helpful:
Gene Expression
Background on proteins: The workhorses of the cell
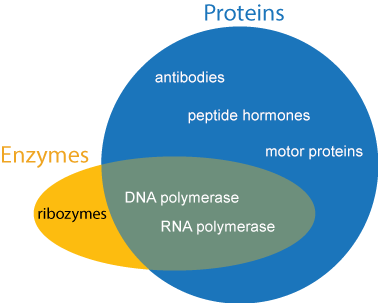
Genes contain the recipes for proteins. Proteins are the true “workhorses” of the cell. Each cell contains an abundance and variety of proteins without which life would be not be possible. Depending on its exact chemical structure and shape, a protein may act as a biological catalyst (called an enzyme), biological signal (such as hormones), energy storage, provide structural integrity to the cell (like the beams in a house), transportation for other compounds (like a taxi or a gate), or have other diverse functions that are essential to life.
Enzymes will be mentioned several times throughout this tutorial, so we will make special mention of them here. Enzymes are compounds that speed up chemical reactions. The words “enzyme” and “protein” are not synonymous. An enzyme is defined by its function and not its chemical makeup, whereas a protein is defined by its chemical makeup and not its function. The vast majority of enzymes are proteins, so when the word “enzyme” is used, you can be almost assured that the enzyme is a protein. However, not all proteins function as enzymes, and not all enzymes are made of protein.
Proteins are made of building blocks called amino acids. Amino acids form chains with each other. These chains are folded in a specific pattern in order to form a precise three-dimensional shape. When an amino acid chain is folded into a three-dimensional shape it is then called a protein. Often, multiple protein subunits (each made from separate amino acid chains) come together to form a protein complex. This partnership is necessary for the protein subunits to complete their true role in the cell. This modular approach to functionality also increases the cell’s flexibility; in some cases, one protein subunit can be swapped out for a slightly different subunit whose function better serves the current situation.

The three-dimensional shape of a protein gives it its identity and helps determine what function it has (enzyme, structural protein, transporter, etc.). In contrast, if amino acid chains are folded incorrectly, the resulting “misfolded” proteins will not perform their intended functions and may instead contribute to the development of diseases, such as Alzheimer’s, Parkinson’s, and Huntington’s.
Proteins are both the end-goal and catalysts of gene expression
Earlier, we said that genes contain the recipes for proteins. Given a little more background, we can now say that a gene contains the instructions for building the particular amino acid chain that is unique to a protein. When a gene is actively signaling a cell to make a certain protein, that gene is said to be expressed. There are three major milestones in gene expression: transcription of DNA to another molecule called RNA, translation to an amino acid chain, and protein folding with modification.

Proteins are the end-product of gene expression, but they also help regulate transcription, translation, and protein folding by acting as enzymes (among other roles). For simplicity, we will describe gene expression as if it were a linear process that begins with DNA and ends with protein. In reality, however, gene expression is part of an enormous, incredibly complex system of feedback loops. Simply put, gene expression produces proteins, which in turn regulate gene expression (sometimes directly and sometimes indirectly).
Transcription
The first step in gene expression is transcription. When a gene is transcribed, its double-stranded DNA is gently unwound by enzymes. The enzymes read the DNA and transcribe the instructions it contains by producing a new molecule called messenger RNA (mRNA; also called an mRNA transcript). The DNA is then rewound.
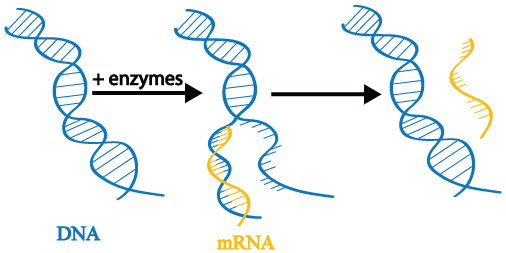
During gene expression, a single gene may be transcribed repeatedly to produce many mRNA copies. As we will see later in this tutorial series, controlling transcription is at the heart of epigenetics.
Translation
Once the DNA has been copied into an mRNA transcript, the instructions in the transcript are then translated into a chain of amino acids by a different enzyme.
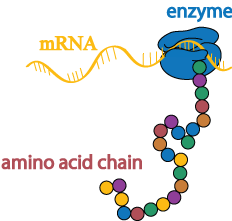
Folding and modifications
Amino acid chains have a natural tendency to fold into three-dimensional proteins, but molecular “chaperones” help ensure that the correct shape is achieved in a timely fashion.

In addition to chaperone-assisted folding, newly-formed proteins may also undergo several kinds of modifications with the help of other molecular machinery. These molecular machines may stabilize the protein’s shape, break off pieces of the protein, or add sugars and other non-protein chemical tags that are meaningful biological signals (these signals may, for example, act like a baggage claim tag, identifying the protein’s final destination inside or outside the cell).
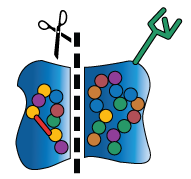
The cell may modify or not modify its proteins in a particular way depending on other conditions within the body (e.g., in response to something as dramatic as a virus or something as mundane as a hormone signal). The ability to add or remove these modifications helps the cell to build a diverse collection of proteins from a finite and (mostly) unchanging DNA code. This added layer of “customization” is another way in which a cell cultivates versatility. While DNA may be the foundation of a biological system, it is in no way the last word on the subject.
Genome Structure
In order to understand epigenetics, it helps to have a broad view of the human genomic landscape. This module will review the basic organization of the genome, from the tiny units called base pairs through larger structures, such as chromosomes.
DNA – A double helix made of base pairs
DNA contains four bases, represented by the letters A (for adenine), T (for thymine), G (for guanine), and C (for cytosine). These bases are attached to a “string” made of a special type of sugar called deoxyribose. DNA contains two sugar strings that are twisted around each other: a double helix. The bases on one side of a double helix pair up with the bases on the other side of the double helix in order to hold the two strings together. The bases in DNA are selective about their pairings. Normally, they will only form two specific combinations: G pairs up with C and A pairs up with T. These A—T and G—C base pairs (abbreviated bp) are not only the foundation of DNA; they are also the basic unit of measurement in the genome.

Figure 12 (above) shows the stereotypical model of DNA: a double-helix shape with color-coded base pairs. While this model provides a good introduction to DNA, it is an oversimplification of the architecture of the human genome. The double-helix model in Figure 1 is like zooming in on a single leaf on a single tree, when what you are really trying to understand is the forest. In reality, there is such a vast amount of DNA in the genome that it must be divided up and condensed many, many times in order to be packaged inside the microscopic cells of the human body. The sections below first quantify exactly how big of a storage problem this is and then slowly zoom out to provide a birds-eye view of the human genome.
Big genomes lead to storage problems
The human genome is very large; it is made of over 6 billion bp,[7]Morton, N. E. Parameters of the human genome. Proc. Natl. Acad. Sci. U. S. A. 88, 7474–6 (1991). or about 2 meters of DNA (over 6 and a half feet),[8]Alberts, B. et al. in Mol. Biol. Cell 191–234 (Garland Science, 2002). at which contains about 38,000 genes. Each cell [9]Ezkurdia, I. et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum. Mol. Genet. (2014). doi:10.1093/hmg/ddu309[10]Bianconi, E. et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 40, 463–71 (2013). in the body contains its own, complete copy of the genome. The massive quantity of DNA in our bodies swiftly causes a storage problem. Consider: the largest cell in the human body is the oocyte, or “egg cell,” which is only found in females and is about one millimeter in diameter — approximately the size of a comma. Additionally, not every cell in the body needs access to all 38,000 genes all the time. If all the cells in every tissue and organ tried to express every gene in the genome simultaneously, the chaos would be completely incompatible with life.
Our bodies manage both the storage problem and the gene expression problem through many levels of DNA compression. Compressing the genome allows it to fit inside the nucleus of a single cell. By controlling which parts of the genome are more or less tightly compressed, a cell can access just those parts of the DNA which are necessary to its long-term role in the body, while retaining the flexibility to respond to pressing demands from a changing environment.

Major motifs in genome compression
DNA compression has three major repeating motifs: lines, coils, and loops.

In the sections that follow, we will watch as a linear strand of DNA becomes coiled (forming chromatosomes). Then the coil itself will also be coiled, looped, and coiled again (forming chromatin). In the end, DNA will be condensed into a shape that looks roughly like a line (forming chromosomes). The contortions that the genome undergoes are dizzying, but it is important to appreciate their complexity in order to understand why epigenetics is a necessary form of regulation in big genomes and how epigenetic changes act on multiple levels of genomic organization.
Beginning with the end in mind: Chromosomes
It will help to know in advance that human DNA does not exist in one continuous double-helix string. Human DNA is divided into 46 “strings” of DNA. These strings can be organized into 23 pairs. Each separate string is called a chromosome, a term which we will circle back around to shortly.

Nucleosomes & chromatosomes
The first level of DNA compression is the nucleosome. Nucleosomes are made of DNA wrapped around a core of proteins. The center of each nucleosome is made of eight pieces of protein called histones, which come together to form a histone core.

If DNA is like a string, then histone cores are the spools about which this string is wound.

Pieces of DNA called linker DNA protrude from each “spool” to connect one nucleosome to the next. A special linker histone makes contact with the linker DNA as it enters and exits the nucleosome. When you add a linker histone to a nucleosome, the entire unit is called a chromatosome.

Each chromatosome can hold only 166 bp of DNA (146 bp around the nucleosome, plus 20 bp held by the linker histone).[11]Bednar, J. & Dimitrov, S. Chromatin under mechanical stress: from single 30 nm fibers to single nucleosomes. FEBS J. 278, 2231–43 (2011).[12]Luger, K., Maeder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8Aa resolution. Nature 389, 251–260 (1997). Human genes vary in length, with a median length of 14,000 bp and an average of 27,000 bp.[13]International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). Thus, many chromatosomes are required to spool up a single gene, and many more are needed to contain the entire genome.

Chromatin
The next level of DNA compression is chromatin. The chromatosome and linker DNA are together twisted into a tight coil called chromatin fiber. The chromatin fiber can be further condensed by looping it and then coiling it again.

Chromosome
The final level of DNA compression is the chromosome. Chromatin fibers are folded on themselves to produce a linear chromosome. Recall: human DNA is not contiguous. It is divided into 46 pieces. Each of these 46 pieces is a separate chromosome, all of which are contained in the nucleus.

Epigenetics
When genes are being expressed (being transcribed), the many levels of DNA compression cause a problem. It is necessary for enzymes to access the DNA and gently pull apart a section of the double helix in order to read the underlying code, but chromatin and histones are a prohibitive barrier. Thus, the cell has ways of modifying histones or temporarily removing them from key sites. Increasing and decreasing the accessibility of specific genes, or even whole chromosomes, is the secret of epigenetics.
Tweaking the degree of compression is one of the major tools of epigenetic regulation.

References
[1] Holliday, R. Epigenetics: A historical overview. Epigenetics 1, 76–80 (2006).
[2] Harper, D. Online Etymology Dictionary. (2014). at http://www.etymonline.com
[3] Pinney, S. E. & Simmons, R. a. Epigenetic mechanisms in the development of type 2 diabetes. Trends Endocrinol. Metab. 21, 223–9 (2010).
[4] Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–59 (2008).
[5] Berger, S. L., Kouzarides, T., Shiekhattar, R. & Shilatifard, A. An operational definition of epigenetics. Genes Dev. 23, 781–3 (2009).
[6] NIH Roadmap Epigenomics. Roadmap Epigenomics Project – Overview. (2010). at http://www.roadmapepigenomics.org/overview
[7]Morton, N. E. Parameters of the human genome. Proc. Natl. Acad. Sci. U. S. A. 88, 7474–6 (1991).
[8] Alberts, B. et al. in Mol. Biol. Cell 191–234 (Garland Science, 2002) at https://www.ncbi.nlm.nih.gov/books/NBK21054/
[9] Ezkurdia, I. et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum. Mol. Genet. (2014). doi:10.1093/hmg/ddu309
[10] Bianconi, E. et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 40, 463–71 (2013).
[11] Bednar, J. & Dimitrov, S. Chromatin under mechanical stress: from single 30 nm fibers to single nucleosomes. FEBS J. 278, 2231–43 (2011).
[12] Luger, K., Maeder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8Aa resolution. Nature 389, 251–260 (1997).
[13] International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Additional Resources
Teaching / Learning Materials
- U. of Utah Genetic Science Learning Center: Epigenetics Multimedia
- National Human Genome Research Institute Fact Sheet: Epigenomics
- Geneimprint: Gateway to gene imprinting information
- EpiGenie Science Writers
- Scitable by Nature Education: Genetics Topic Room
Organizations and Landmark Projects
- International Human Epigenome Consortium
- NIH Roadmap Epigenomics Project
- ENCODE: Encyclopedia of DNA Elements
- NHGRI: National Human Genome Research Institute
- NIH Epigenomics Program
Scholarly Review Articles
- Enivronmental epigenomics and disease susceptibility Jirtle R. and Skinner M., Nat Rev Genet (2007) 8(4):253-62 doi: 10.1038/nrg2045
- Epigenetics, chromatin and genome organization: recent advances from the ENCODE project Siggens L. and Ekwall K., J Intern Med (2014) doi: 10.1111/joim.12231
- Epigenetics as a unifying principle in the aetiology of complex traits and diseases Petronis A., Nature (2010) 465(7299):721-7 doi: 10.1038/nature09230
- Epigenetics and the environmental regulation of the genome and its function Zhang T. and Meaney M., Annu Rev Psychol (2010) 61:439-66 doi: 10.1146/annurev.psych.60.110707.163625